Create a robustness or sequential plot
PlotRobustnessSequential.RdCreate a robustness or sequential plot
PlotRobustnessSequential(
dfLines,
dfPoints = NULL,
BF = NULL,
hasRightAxis = TRUE,
xName = NULL,
yName = NULL,
addEvidenceArrowText = TRUE,
drawPizzaTxt = !is.null(BF),
evidenceLeveltxt = !is.null(BF),
pointLegend = !is.null(dfPoints),
linesLegend = !is.null(dfLines$g),
bfSubscripts = NULL,
pizzaTxt = hypothesis2BFtxt(hypothesis)$pizzaTxt,
bfType = c("BF01", "BF10", "LogBF10"),
hypothesis = c("equal", "smaller", "greater"),
pointColors = c("red", "grey", "black", "white"),
lineColors = c("black", "grey", "black"),
lineTypes = c("solid", "solid", "dotted"),
addLineAtOne = TRUE,
bty = list(type = "n", ldwX = 0.5, lwdY = 0.5),
plotLineOrPoint = c("auto", "line", "point"),
pointShape = rep(21, 3),
pointFill = c("grey", "black", "white"),
pointColor = rep("black", 3),
pointSize = c(3, 2, 2),
evidenceTxt = NULL,
arrowLabel = NULL,
...
)Arguments
- dfLines
A dataframe with
$x,$y, and optionally$g.$yis assumed to be on the log scale.- dfPoints
A dataframe with
$x,$y, and optionally$g.- BF
Numeric, with value of Bayes factor. This MUST correspond to bfType.
- hasRightAxis
Logical, should there be a right axis displaying evidence?
- xName
String or expression, displayed on the x-axis.
- yName
String or expression, displayed on the y-axis.
- addEvidenceArrowText
Logical, should arrows indicating "Evidence for H0/H1" be drawn?
- drawPizzaTxt
Logical, should there be text above and below the pizza plot?
- evidenceLeveltxt
Logical, should "Evidence for H0: extreme" be drawn? Ignored if
!is.null(dfLines$g) && linesLegend.- pointLegend
Logical, should a legend of
dfPoints$gbe shown?- linesLegend
Logical, should a legend of
dfLines$gbe shown?- bfSubscripts
String, manually specify the BF labels.
- pizzaTxt
String vector of length 2, text to be drawn above and below pizza plot.
- bfType
String, what is the type of BF? Options are "BF01", "BF10", or "LogBF10".
- hypothesis
String, what was the hypothesis? Options are "equal", "smaller", or "greater".
- pointColors
String vector, colors for points if
dfPoints$gis notNULL.- lineColors
String vector, colors for lines if
dfLines$gis notNULL.- lineTypes
String vector, line types if
dfLines$gis notNULL.- addLineAtOne
Logical, should a black line be draw at BF = 1?
- bty
List of three elements. Type specifies the box type, ldwX the width of the x-axis, lwdY the width of the y-axis.
- plotLineOrPoint
String, should the main geom in the plot be a line or a point? If set to auto, points are shown whenever
nrow(dfLines) <= 60.- pointShape
String, if
plotLineOrPoint == "point"then this controls the shape aesthetic.- pointFill
String, if
plotLineOrPoint == "point"then this controls the fill aesthetic.- pointColor
String, if
plotLineOrPoint == "point"then this controls the color aesthetic.- pointSize
String, if
plotLineOrPoint == "point"then this controls the size aesthetic.- evidenceTxt
String to display evidence level in the topright of the plot. If NULL then a default is used.
- arrowLabel
String to display text next to arrows inside the plot. If NULL then a default is used.
- ...
Unused.
Examples
# Data for sequential
#
set.seed(1)
n <- 100
dfLines <- data.frame(
x = seq_len(n),
y = c(0, rnorm(n - 1, seq_len(n - 1) / 30, .5)) # log Bayes factor
)
# Sequential plot
#
PlotRobustnessSequential(
dfLines = dfLines,
xName = "n",
)
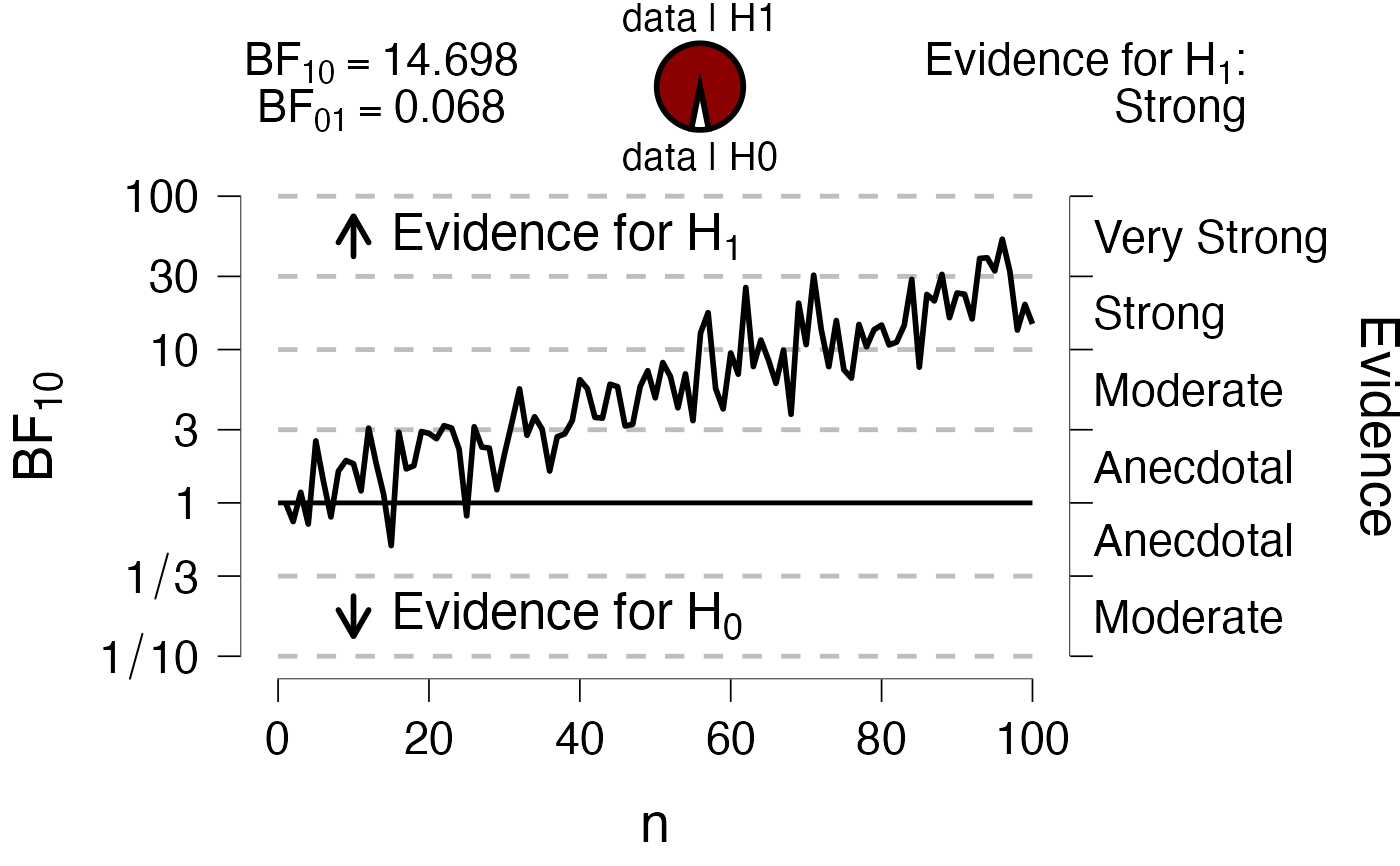
 # Sequential plot add info
#
BF10 <- exp(tail(dfLines, 1)$y)
PlotRobustnessSequential(
dfLines = dfLines,
xName = "n",
BF = BF10,
bfType = "BF10"
)
# Sequential plot add info
#
BF10 <- exp(tail(dfLines, 1)$y)
PlotRobustnessSequential(
dfLines = dfLines,
xName = "n",
BF = BF10,
bfType = "BF10"
)
 # Data for robustness plot
n <- 100
x <- seq_len(n)/100
y <- cos(pi*x)
dfLines <- data.frame(
x = x,
y = y
)
# Robustness plot
#
PlotRobustnessSequential(
dfLines = dfLines,
xName = "Prior width"
)
# Data for robustness plot
n <- 100
x <- seq_len(n)/100
y <- cos(pi*x)
dfLines <- data.frame(
x = x,
y = y
)
# Robustness plot
#
PlotRobustnessSequential(
dfLines = dfLines,
xName = "Prior width"
)
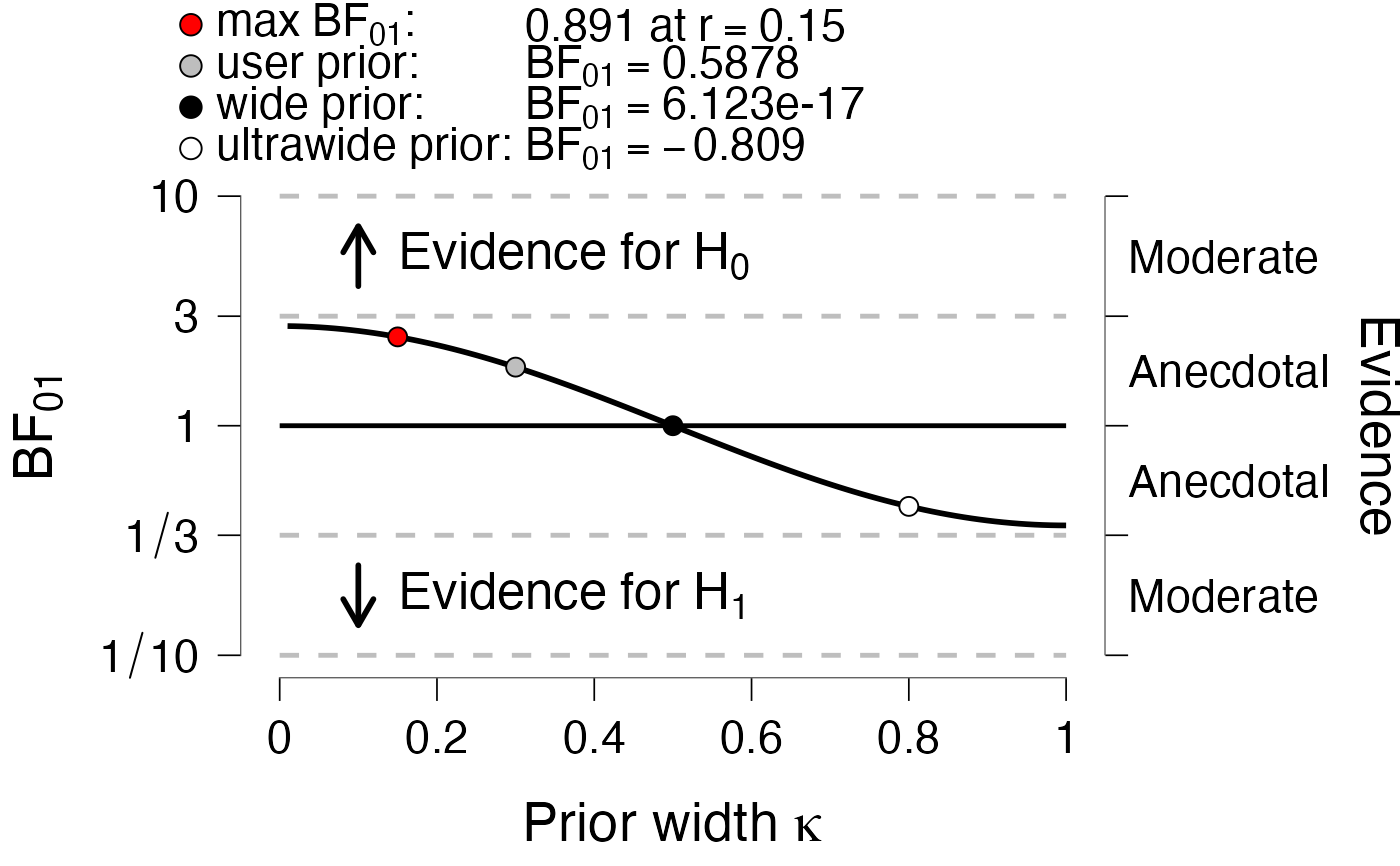
 # In JASP, we often prefer more information in the plot
x <- dfLines$x[c(15, 30, 50, 80)]
y <- dfLines$y[c(15, 30, 50, 80)]
maxBFrVal <- x[1]
maxBF10 <- y[1]
BF10user <- y[2]
BF10w <- y[3]
BF10ultra <- y[4]
BFsubscript <- "[0][1]"
label1 <- c(
gettextf("max BF%s", BFsubscript),
gettext("user prior"),
gettext("wide prior"),
gettext("ultrawide prior")
)
# some failsafes to parse translations as expressions
label1[1] <- gsub(pattern = "\\s+", "~", label1[1])
label1[-1] <- paste0("\"", label1[-1], "\"")
label1 <- paste0("paste(", label1, ", ':')")
BFandSubscript <- gettextf("BF%s", BFsubscript)
BFandSubscript <- gsub(pattern = "\\s+", "~", BFandSubscript)
label2 <- c(
gettextf("%s at r==%s", format(maxBF10, digits = 4), format(maxBFrVal, digits = 4)),
paste0(BFandSubscript, "==", format(BF10user, digits = 4)),
paste0(BFandSubscript, "==", format(BF10w, digits = 4)),
paste0(BFandSubscript, "==", format(BF10ultra,digits = 4))
)
label2[1L] <- gsub(pattern = "\\s+", "~", label2[1])
dfPoints <- data.frame(
x = x,
y = c(maxBF10, BF10user, BF10w, BF10ultra),
g = label1,
label1 = jaspGraphs::parseThis(label1),
label2 = jaspGraphs::parseThis(label2),
stringsAsFactors = FALSE
)
PlotRobustnessSequential(
dfLines = dfLines,
xName = expression(paste("Prior width ", kappa)),
dfPoints = dfPoints
)
# In JASP, we often prefer more information in the plot
x <- dfLines$x[c(15, 30, 50, 80)]
y <- dfLines$y[c(15, 30, 50, 80)]
maxBFrVal <- x[1]
maxBF10 <- y[1]
BF10user <- y[2]
BF10w <- y[3]
BF10ultra <- y[4]
BFsubscript <- "[0][1]"
label1 <- c(
gettextf("max BF%s", BFsubscript),
gettext("user prior"),
gettext("wide prior"),
gettext("ultrawide prior")
)
# some failsafes to parse translations as expressions
label1[1] <- gsub(pattern = "\\s+", "~", label1[1])
label1[-1] <- paste0("\"", label1[-1], "\"")
label1 <- paste0("paste(", label1, ", ':')")
BFandSubscript <- gettextf("BF%s", BFsubscript)
BFandSubscript <- gsub(pattern = "\\s+", "~", BFandSubscript)
label2 <- c(
gettextf("%s at r==%s", format(maxBF10, digits = 4), format(maxBFrVal, digits = 4)),
paste0(BFandSubscript, "==", format(BF10user, digits = 4)),
paste0(BFandSubscript, "==", format(BF10w, digits = 4)),
paste0(BFandSubscript, "==", format(BF10ultra,digits = 4))
)
label2[1L] <- gsub(pattern = "\\s+", "~", label2[1])
dfPoints <- data.frame(
x = x,
y = c(maxBF10, BF10user, BF10w, BF10ultra),
g = label1,
label1 = jaspGraphs::parseThis(label1),
label2 = jaspGraphs::parseThis(label2),
stringsAsFactors = FALSE
)
PlotRobustnessSequential(
dfLines = dfLines,
xName = expression(paste("Prior width ", kappa)),
dfPoints = dfPoints
)
 # convenience function for showing plots side by side. You may need to click zoom in Rstudio
# to properly view the plots.
showSideBySide <- function(..., nrow = 1L, ncol = ...length()) {
require(gridExtra)
jaspgraphplot2grob <- function(x) {
if (!inherits(x, "jaspGraphsPlot")) return(x)
else return(x$plotFunction(x$subplots, args = x$plotArgs, grob = TRUE))
}
gridExtra::grid.arrange(gridExtra::arrangeGrob(
grobs = lapply(list(...), jaspgraphplot2grob), nrow = nrow, ncol = ncol))
}
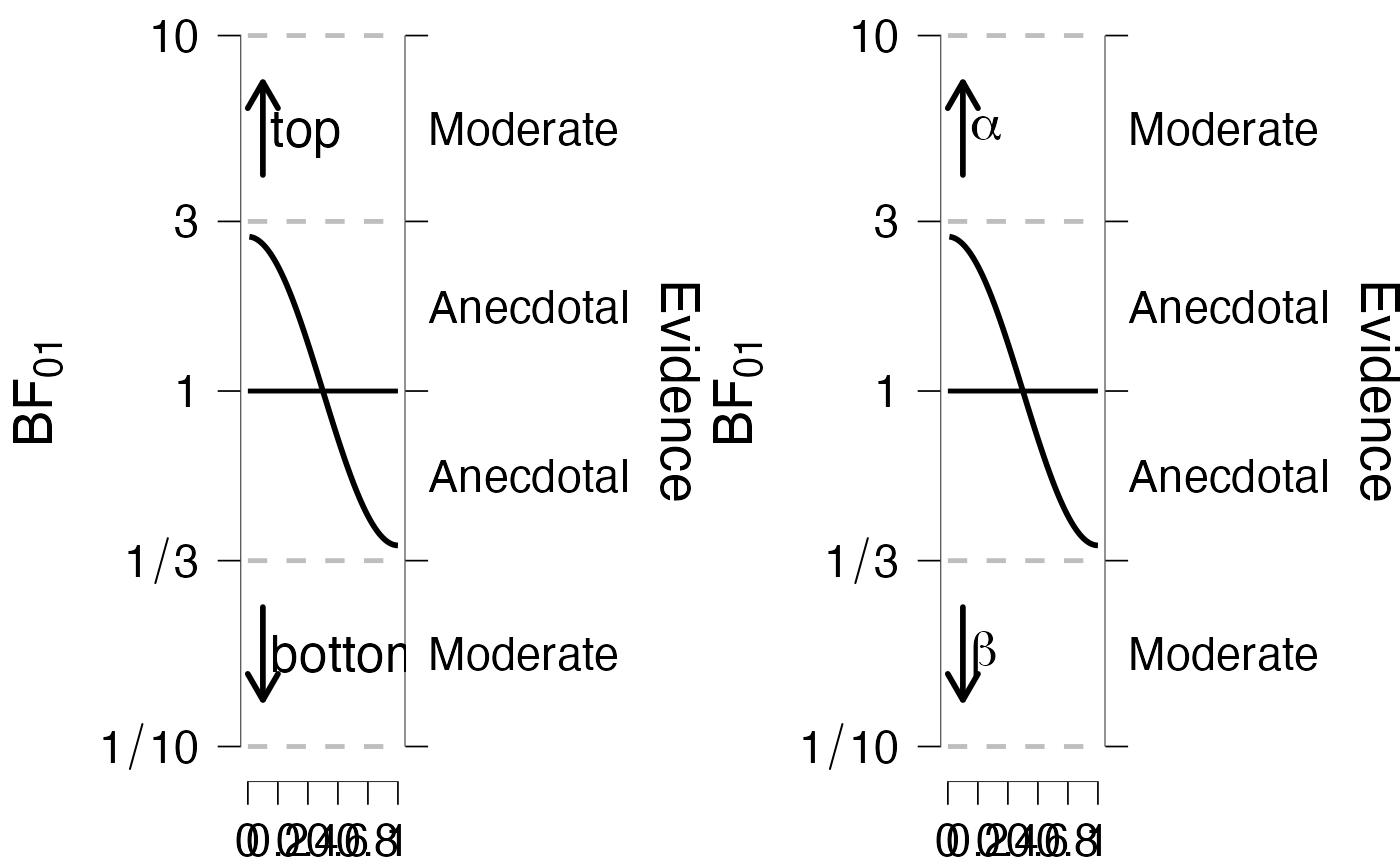
# arrow labels can be modified
g1 <- PlotRobustnessSequential(dfLines = dfLines, arrowLabel = c("top", "bottom"))
g2 <- PlotRobustnessSequential(dfLines = dfLines,
arrowLabel = jaspGraphs::parseThis(c("alpha", "beta")))
showSideBySide(g1, g2)
#> Loading required package: gridExtra
# convenience function for showing plots side by side. You may need to click zoom in Rstudio
# to properly view the plots.
showSideBySide <- function(..., nrow = 1L, ncol = ...length()) {
require(gridExtra)
jaspgraphplot2grob <- function(x) {
if (!inherits(x, "jaspGraphsPlot")) return(x)
else return(x$plotFunction(x$subplots, args = x$plotArgs, grob = TRUE))
}
gridExtra::grid.arrange(gridExtra::arrangeGrob(
grobs = lapply(list(...), jaspgraphplot2grob), nrow = nrow, ncol = ncol))
}
# arrow labels can be modified
g1 <- PlotRobustnessSequential(dfLines = dfLines, arrowLabel = c("top", "bottom"))
g2 <- PlotRobustnessSequential(dfLines = dfLines,
arrowLabel = jaspGraphs::parseThis(c("alpha", "beta")))
showSideBySide(g1, g2)
#> Loading required package: gridExtra
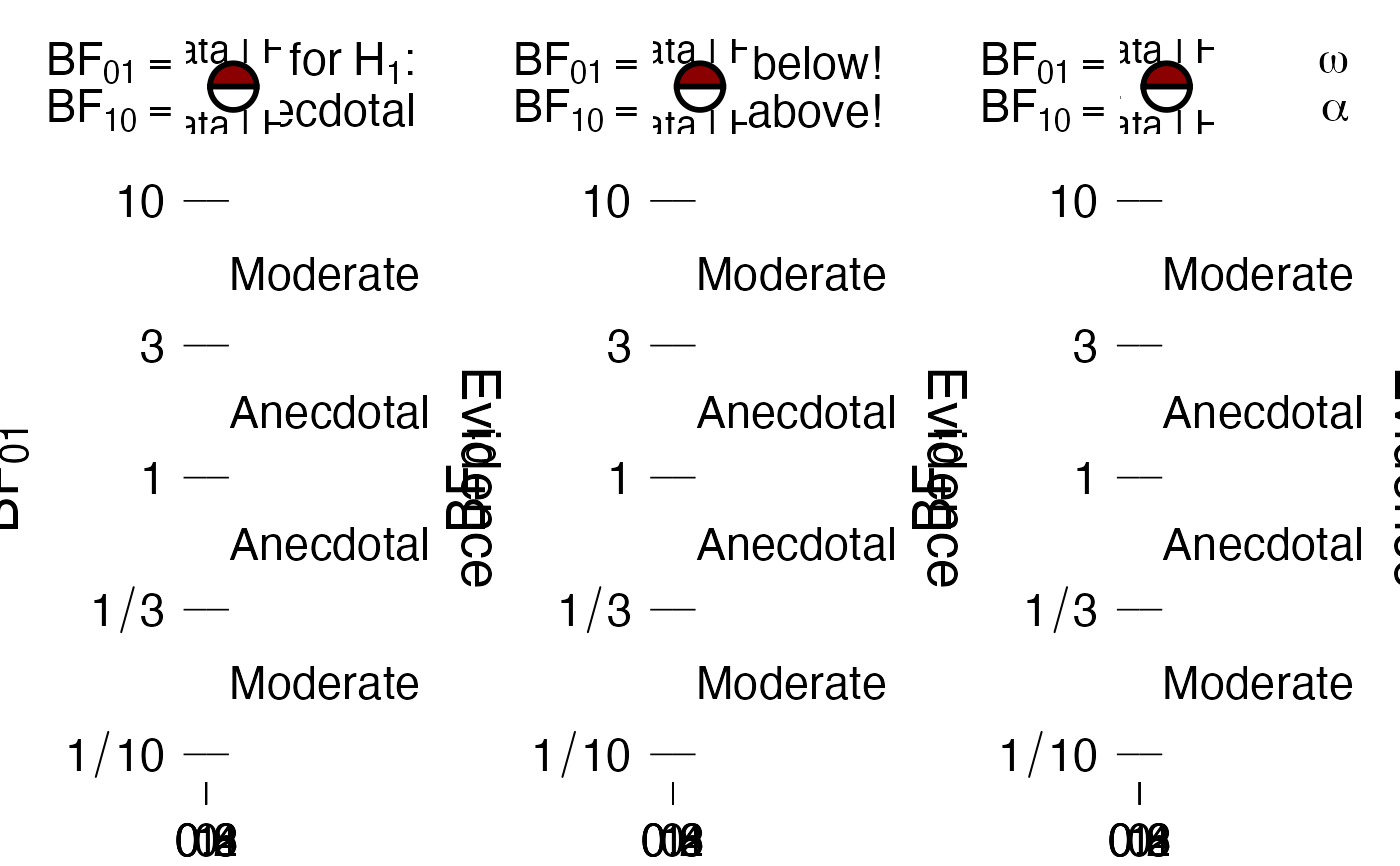
 # text in the top right (evidence text) can be modified
g1 <- PlotRobustnessSequential(dfLines = dfLines, BF = 1)
g2 <- PlotRobustnessSequential(dfLines = dfLines, BF = 1,
evidenceTxt = c("I'm above!", "I'm below!"))
g3 <- PlotRobustnessSequential(dfLines = dfLines, BF = 1,
evidenceTxt = jaspGraphs::parseThis(c("alpha", "omega")))
showSideBySide(g1, g2, g3)
# text in the top right (evidence text) can be modified
g1 <- PlotRobustnessSequential(dfLines = dfLines, BF = 1)
g2 <- PlotRobustnessSequential(dfLines = dfLines, BF = 1,
evidenceTxt = c("I'm above!", "I'm below!"))
g3 <- PlotRobustnessSequential(dfLines = dfLines, BF = 1,
evidenceTxt = jaspGraphs::parseThis(c("alpha", "omega")))
showSideBySide(g1, g2, g3)